Chromosome 3
| Chromosome 3 | |
|---|---|
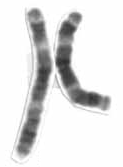 Human chromosome 3 pair after G-banding. One is from mother, one is from father. | |
 Chromosome 3 pair in human male karyogram. | |
| Features | |
| Length (bp) | 198,295,559 bp (GRCh38)[1] |
No. of genes | 1,024 (CCDS)[2] |
| Type | Autosome |
| Centromere position | Metacentric[3] (90.9 Mbp[4]) |
| Complete gene lists | |
| CCDS | Gene list |
| HGNC | Gene list |
| UniProt | Gene list |
| NCBI | Gene list |
| External map viewers | |
| Ensembl | Chromosome 3 |
| Entrez | Chromosome 3 |
| NCBI | Chromosome 3 |
| UCSC | Chromosome 3 |
| Full DNA sequences | |
| RefSeq | NC_000003 (FASTA) |
| GenBank | CM000665 (FASTA) |
Chromosome 3 is one of the 23 pairs of chromosomes in humans. People normally have two copies of this chromosome. Chromosome 3 spans almost 200 million base pairs (the building material of DNA) and represents about 6.5 percent of the total DNA in cells.
Contents
1 Genes
1.1 Number of genes
1.2 List of genes
1.2.1 p-arm
1.2.2 q-arm
2 Diseases and disorders
3 Cytogenetic band
4 References
5 External links
Genes
Number of genes
The following are some of the gene count estimates of human chromosome 3. Because researchers use different approaches to genome annotation their predictions of the number of genes on each chromosome varies (for technical details, see gene prediction). Among various projects, the collaborative consensus coding sequence project (CCDS) takes an extremely conservative strategy. So CCDS's gene number prediction represents a lower bound on the total number of human protein-coding genes.[5]
| Estimated by | Protein-coding genes | Non-coding RNA genes | Pseudogenes | Source | Release date |
|---|---|---|---|---|---|
| CCDS | 1,024 | — | — | [2] | 2016-09-08 |
| HGNC | 1,036 | 483 | 761 | [6] | 2017-05-12 |
| Ensembl | 1,073 | 1,158 | 761 | [7] | 2017-03-29 |
| UniProt | 1,081 | — | — | [8] | 2018-02-28 |
| NCBI | 1,085 | 1,108 | 902 | [9][10][11] | 2017-05-19 |
List of genes
The following is a partial list of genes on human chromosome 3. For complete list, see the link in the infobox on the right.
p-arm
Partial list of the genes located on p-arm (short arm) of human chromosome 3:
ALAS1: aminolevulinate, delta-, synthase 1
APEH: encoding enzyme Acylamino-acid-releasing enzyme
ARPP-21: Cyclic AMP-regulated phosphoprotein, 21 kDa
AZI2: encoding protein 5-azacytidine-induced protein 2
BRK1: SCAR/WAVE actin nucleating complex subunit
BRPF1: bromodomain and PHD finger containing 1
BTD: biotinidase
C3orf14-Chromosome 3 open reading frame 14: predicted DNA binding protein.
C3orf23: encoding protein Uncharacterized protein C3orf23
C3orf60/NDUFAF3: encoding enzyme NADH dehydrogenase [ubiquinone] 1 alpha subcomplex assembly factor 3
C3orf62: chromosome 3 open reading frame 62
CACNA2D3: calcium channel, voltage-dependent, alpha 2/delta subunit 3
CCR5: chemokine (C-C motif) receptor 5
CGGBP1: CGG triplet repeat binding protein 1
CMTM7: CKLF like MARVEL transmembrane domain containing 7
CNTN4: Contactin 4
COL7A1: Collagen, type VII, alpha 1 (epidermolysis bullosa, dystrophic, dominant and recessive)
CRBN: Cereblon protein[12]
DCLK3: Doublecortin like kinase 3
EAF1: ELL associated factor 1
ENTPD3: ectonucleoside triphosphate diphosphohydrolase 3
FAM107A: Family with sequence similarity 107 member A
FAM19A1: Family with sequence similarity 19 member A1, C-C motif chemokine like
FBXL2: F-box and leucine rich repeat protein 2
FOXP1: Forkhead Box Protein P1
FRA3A encoding protein Fragile site, aphidicolin type, common, fra(3)(p24.2)
FRMD4B encoding protein FERM domain containing 4B
GMPPB: GDP-mannose pyrophosphorylase B
HEMK1: encoding protein HemK methyltransferase family member 1
HIGD1A: HIG1 domain family member 1A
LARS2: leucyl-tRNA synthetase, mitochondrial
LIMD1: LIM domain-containing protein 1
LINC00312: Long intergenic non-protein-coding RNA 312
MITF: microphthalmia-associated transcription factor
MLH1: mutL homolog 1, colon cancer, nonpolyposis type 2 (E. coli)
MYRIP: Myosin VIIA and Rab interacting protein
NBEAL2: Neurobeachin-like 2
NKTR: NK-tumor recognition protein
NPRL2: Nitrogen permease regulator 2-like protein
OXTR: oxytocin receptor
PHF7 encoding protein PHD finger protein 7
PTHR1: parathyroid hormone receptor 1
QRICH1: encoding protein QRICH1, also known as Glutamine-rich protein 1,
RBM6: RNA-binding protein 6
RPP14: Ribonuclease P protein subunit p14
SCN5A: sodium channel, voltage-gated, type V, alpha (long QT syndrome 3)
SETD5: SET domain containing 5
SFMBT1: Scm-like with four mbt domains 1
SLC25A20: solute carrier family 25 (carnitine/acylcarnitine translocase), member 20
STT3B: catalytic subunit of the oligosaccharyltransferase complex
SYNPR: synaptoporin
TDGF1: Teratocarcinoma-derived growth factor 1
TMEM158: Transmembrane protein 158
TMIE: transmembrane inner ear
TRAK1: trafficking kinesin-binding protein 1
TRANK1: encoding protein Tetratricopeptide repeat and ankyrin repeat containing 1
TUSC2: tumor suppressor candidate 2
UCN2: Urocortin-2
ULK4: UNC-51 like kinase 4
VGLL3: vestigial-like family member 3
VHL: von Hippel-Lindau tumor suppressor
ZMYND10: zinc finger MYND-type containing 10
ZNF502: encoding protein Zinc finger protein 502
ZNF621: encoding protein Zinc finger protein 621
q-arm
Partial list of the genes located on q-arm (long arm) of human chromosome 3:
ADIPOQ: adiponectin
AMOTL2: encoding protein Angiomotin-like protein 2
ARHGAP31: Rho GRPase activating protein 31
C3orf1: chromosome 3 open reading frame 1
C3orf70 chromosome 3 open reading frame 70
CAMPD1: Camptodactyly
CCDC80: Coiled-coil domain containing protein 80
CD200R1: Cell surface glycoprotein CD200 receptor 1
CLDND1: Claudin domain containing 1
CPN2: Carboxypeptidase N subunit 2
CPOX: coproporphyrinogen oxidase (coproporphyria, harderoporphyria)
DPPA2: Developmental pluripotency associated 2
DZIP3 encoding protein DAZ interacting zinc finger protein 3
EAF2: ELL associated factor 2
EFCC1: EF-hand and coiled-coil domain containing 1
ETM1: Essential tremor 1
ETV5: ETS variant 5
FAM43A: family with sequence similarity 43 member A
FAM162A: family with sequence similarity 162 member A
GYG1: Glycogenin-1
HACD2 encoding protein 3-hydroxyacyl-CoA dehydratase 2
HGD: homogentisate 1,2-dioxygenase (homogentisate oxidase)
IFT122: intraflagellar transport gene 122
KIAA1257: KIAA1257
LMLN: encoding protein Leishmanolysin-like (metallopeptidase M8 family)
LRRC15: leucine rich repeat containing 15
LSG1: large subunit GTPase 1 homolog
MB21D2: encoding protein Mab-21 domain containing 2
MCCC1: methylcrotonoyl-Coenzyme A carboxylase 1 (alpha)
MYLK: Telokin
NFKBIZ: NF-kappa-B inhibitor zeta
PARP14 encoding protein Poly(ADP-ribose) polymerase family member 14
PCCB: propionyl Coenzyme A carboxylase, beta polypeptide
PDCD10: programmed cell death 10
PIK3CA: phosphoinositide-3-kinase, catalytic, alpha polypeptide
PROSER1: Proline and serine rich protein 1
RAB7: RAB7, member RAS oncogene family
RETNLB: resistin-like beta
RHO: rhodopsin visual pigment
RIOX2: Ribosomal oxygenase 2
SELT: Selenoprotein T
SENP7: Sentrin-specific protease 7
SERP1: Stress-associated endoplasmic reticulum protein 1
SOX2: transcription factor
SOX2OT: SOX2 overlapping transcript
SPG14 encoding protein Spastic paraplegia 14 (autosomal recessive)
SRPRB: Signal recognition particle receptor subunit beta
TM4SF1: Transmembrane 4 L6 family member 1
TRAT1: T-cell receptor-associated transmembrane adapter 1
USH3A: Usher syndrome 3A
ZBED2: encoding protein Zinc finger BED-type containing 2
ZNF9: zinc finger protein 9 (a cellular retroviral nucleic acid binding protein)
Diseases and disorders
The following diseases and disorders are some of those related to genes on chromosome 3:
- 3-methylcrotonyl-CoA carboxylase deficiency
- 3q29 microdeletion syndrome
Acute Myeloid Leukemia (AML)- Alkaptonuria
- Arrhythmogenic right ventricular dysplasia
- Atransferrinemia
- Autism
- Autosomal Dominant Optic Atrophy
- ADOA Plus Syndrome
- Biotinidase deficiency
- Blepharophimosis, epicanthus inversus and ptosis type 1
- Breast/colon/lung/pancreatic cancer
- Brugada syndrome
- Castillo fever
- Carnitine-acylcarnitine translocase deficiency
- Cataracts
- Cerebral cavernous malformation
- Charcot-Marie-Tooth disease, type 2
- Charcot-Marie-Tooth disease
- Chromosome 3q duplication syndrome
- Coproporphyria
- Dandy-Walker syndrome
- Deafness
- Diabetes
- Dystrophic epidermolysis bullosa
- Endplate acetylcholinesterase deficiency
- Essential tremors
- Ectrodactyly, Case 4
Glaucoma, primary open angle- Glycogen storage disease
- Hailey-Hailey disease
- Harderoporphyrinuria
- Heart block, progressive/nonprogressive
- Hereditary coproporphyria
- Hereditary nonpolyposis colorectal cancer
HIV infection, susceptibility/resistance to- Hypobetalipoproteinemia, familial
- Hypothermia
- Leukoencephalopathy with vanishing white matter
- Long QT syndrome
- Lymphomas
- Malignant hyperthermia susceptibility
- Metaphyseal chondrodysplasia, Murk Jansen type
- Microcoria
- Moebius syndrome
- Moyamoya disease
- Mucopolysaccharidosis
- Muir-Torre family cancer syndrome
- Myotonic dystrophy
- Neuropathy, hereditary motor and sensory, Okinawa type
- Night blindness
- Nonsyndromic deafness
- Ovarian cancer
- Porphyria
- Propionic acidemia
- Protein S deficiency
- Pseudo-Zellweger syndrome
- Retinitis pigmentosa
- Romano-Ward syndrome
- Seckel Syndrome
- Sensenbrenner syndrome
- Septo-optic dysplasia
- Short stature
- Spinocerebellar ataxia
- Sucrose intolerance
- T-cell leukemia translocation altered gene
- Usher syndrome
- von Hippel-Lindau syndrome
- Waardenburg syndrome
- Xeroderma pigmentosum, complementation group c
Cytogenetic band
.mw-parser-output .tmulti .thumbinner{display:flex;flex-direction:column}.mw-parser-output .tmulti .trow{display:flex;flex-direction:row;clear:left;flex-wrap:wrap;width:100%;box-sizing:border-box}.mw-parser-output .tmulti .tsingle{margin:1px;float:left}.mw-parser-output .tmulti .theader{clear:both;font-weight:bold;text-align:center;align-self:center;background-color:transparent;width:100%}.mw-parser-output .tmulti .thumbcaption{text-align:left;background-color:transparent}.mw-parser-output .tmulti .text-align-left{text-align:left}.mw-parser-output .tmulti .text-align-right{text-align:right}.mw-parser-output .tmulti .text-align-center{text-align:center}@media all and (max-width:720px){.mw-parser-output .tmulti .thumbinner{width:100%!important;box-sizing:border-box;max-width:none!important;align-items:center}.mw-parser-output .tmulti .trow{justify-content:center}.mw-parser-output .tmulti .tsingle{float:none!important;max-width:100%!important;box-sizing:border-box;text-align:center}.mw-parser-output .tmulti .thumbcaption{text-align:center}}
| Chr. | Arm[17] | Band[18] | ISCN start[19] | ISCN stop[19] | Basepair start | Basepair stop | Stain[20] | Density |
|---|---|---|---|---|---|---|---|---|
| 3 | p | 26.3 | 0 | 175 | 7000100000000000000♠1 | 7006280000000000000♠2,800,000 | gpos | 50 |
| 3 | p | 26.2 | 175 | 263 | 7006280000100000000♠2,800,001 | 7006400000000000000♠4,000,000 | gneg | |
| 3 | p | 26.1 | 263 | 408 | 7006400000100000000♠4,000,001 | 7006810000000000000♠8,100,000 | gpos | 50 |
| 3 | p | 25.3 | 408 | 642 | 7006810000100000000♠8,100,001 | 7007116000000000000♠11,600,000 | gneg | |
| 3 | p | 25.2 | 642 | 759 | 7007116000010000000♠11,600,001 | 7007132000000000000♠13,200,000 | gpos | 25 |
| 3 | p | 25.1 | 759 | 963 | 7007132000010000000♠13,200,001 | 7007163000000000000♠16,300,000 | gneg | |
| 3 | p | 24.3 | 963 | 1269 | 7007163000010000000♠16,300,001 | 7007238000000000000♠23,800,000 | gpos | 100 |
| 3 | p | 24.2 | 1269 | 1357 | 7007238000010000000♠23,800,001 | 7007263000000000000♠26,300,000 | gneg | |
| 3 | p | 24.1 | 1357 | 1561 | 7007263000010000000♠26,300,001 | 7007308000000000000♠30,800,000 | gpos | 75 |
| 3 | p | 23 | 1561 | 1751 | 7007308000010000000♠30,800,001 | 7007320000000000000♠32,000,000 | gneg | |
| 3 | p | 22.3 | 1751 | 1926 | 7007320000010000000♠32,000,001 | 7007364000000000000♠36,400,000 | gpos | 50 |
| 3 | p | 22.2 | 1926 | 2013 | 7007364000010000000♠36,400,001 | 7007393000000000000♠39,300,000 | gneg | |
| 3 | p | 22.1 | 2013 | 2188 | 7007393000010000000♠39,300,001 | 7007436000000000000♠43,600,000 | gpos | 75 |
| 3 | p | 21.33 | 2188 | 2451 | 7007436000010000000♠43,600,001 | 7007441000000000000♠44,100,000 | gneg | |
| 3 | p | 21.32 | 2451 | 2626 | 7007441000010000000♠44,100,001 | 7007442000000000000♠44,200,000 | gpos | 50 |
| 3 | p | 21.31 | 2626 | 3239 | 7007442000010000000♠44,200,001 | 7007506000000000000♠50,600,000 | gneg | |
| 3 | p | 21.2 | 3239 | 3385 | 7007506000010000000♠50,600,001 | 7007523000000000000♠52,300,000 | gpos | 25 |
| 3 | p | 21.1 | 3385 | 3676 | 7007523000010000000♠52,300,001 | 7007544000000000000♠54,400,000 | gneg | |
| 3 | p | 14.3 | 3676 | 3910 | 7007544000010000000♠54,400,001 | 7007586000000000000♠58,600,000 | gpos | 50 |
| 3 | p | 14.2 | 3910 | 4143 | 7007586000010000000♠58,600,001 | 7007638000000000000♠63,800,000 | gneg | |
| 3 | p | 14.1 | 4143 | 4362 | 7007638000010000000♠63,800,001 | 7007697000000000000♠69,700,000 | gpos | 50 |
| 3 | p | 13 | 4362 | 4566 | 7007697000010000000♠69,700,001 | 7007741000000000000♠74,100,000 | gneg | |
| 3 | p | 12.3 | 4566 | 4814 | 7007741000010000000♠74,100,001 | 7007798000000000000♠79,800,000 | gpos | 75 |
| 3 | p | 12.2 | 4814 | 4946 | 7007798000010000000♠79,800,001 | 7007835000000000000♠83,500,000 | gneg | |
| 3 | p | 12.1 | 4946 | 5077 | 7007835000010000000♠83,500,001 | 7007871000000000000♠87,100,000 | gpos | 75 |
| 3 | p | 11.2 | 5077 | 5135 | 7007871000010000000♠87,100,001 | 7007878000000000000♠87,800,000 | gneg | |
| 3 | p | 11.1 | 5135 | 5266 | 7007878000010000000♠87,800,001 | 7007909000000000000♠90,900,000 | acen | |
| 3 | q | 11.1 | 5266 | 5427 | 7007909000010000000♠90,900,001 | 7007940000000000000♠94,000,000 | acen | |
| 3 | q | 11.2 | 5427 | 5602 | 7007940000010000000♠94,000,001 | 7007986000000000000♠98,600,000 | gvar | |
| 3 | q | 12.1 | 5602 | 5762 | 7007986000010000000♠98,600,001 | 7008100300000000000♠100,300,000 | gneg | |
| 3 | q | 12.2 | 5762 | 5850 | 7008100300001000000♠100,300,001 | 7008101200000000000♠101,200,000 | gpos | 25 |
| 3 | q | 12.3 | 5850 | 5996 | 7008101200001000000♠101,200,001 | 7008103100000000000♠103,100,000 | gneg | |
| 3 | q | 13.11 | 5996 | 6229 | 7008103100001000000♠103,100,001 | 7008106500000000000♠106,500,000 | gpos | 75 |
| 3 | q | 13.12 | 6229 | 6361 | 7008106500001000000♠106,500,001 | 7008108200000000000♠108,200,000 | gneg | |
| 3 | q | 13.13 | 6361 | 6594 | 7008108200001000000♠108,200,001 | 7008111600000000000♠111,600,000 | gpos | 50 |
| 3 | q | 13.2 | 6594 | 6682 | 7008111600001000000♠111,600,001 | 7008113700000000000♠113,700,000 | gneg | |
| 3 | q | 13.31 | 6682 | 6871 | 7008113700001000000♠113,700,001 | 7008117600000000000♠117,600,000 | gpos | 75 |
| 3 | q | 13.32 | 6871 | 6973 | 7008117600001000000♠117,600,001 | 7008119300000000000♠119,300,000 | gneg | |
| 3 | q | 13.33 | 6973 | 7148 | 7008119300001000000♠119,300,001 | 7008122200000000000♠122,200,000 | gpos | 75 |
| 3 | q | 21.1 | 7148 | 7294 | 7008122200001000000♠122,200,001 | 7008124100000000000♠124,100,000 | gneg | |
| 3 | q | 21.2 | 7294 | 7440 | 7008124100001000000♠124,100,001 | 7008126100000000000♠126,100,000 | gpos | 25 |
| 3 | q | 21.3 | 7440 | 7674 | 7008126100001000000♠126,100,001 | 7008129500000000000♠129,500,000 | gneg | |
| 3 | q | 22.1 | 7674 | 7936 | 7008129500001000000♠129,500,001 | 7008134000000000000♠134,000,000 | gpos | 25 |
| 3 | q | 22.2 | 7936 | 8053 | 7008134000001000000♠134,000,001 | 7008136000000000000♠136,000,000 | gneg | |
| 3 | q | 22.3 | 8053 | 8228 | 7008136000001000000♠136,000,001 | 7008139000000000000♠139,000,000 | gpos | 25 |
| 3 | q | 23 | 8228 | 8461 | 7008139000001000000♠139,000,001 | 7008143100000000000♠143,100,000 | gneg | |
| 3 | q | 24 | 8461 | 8811 | 7008143100001000000♠143,100,001 | 7008149200000000000♠149,200,000 | gpos | 100 |
| 3 | q | 25.1 | 8811 | 9001 | 7008149200001000000♠149,200,001 | 7008152300000000000♠152,300,000 | gneg | |
| 3 | q | 25.2 | 9001 | 9162 | 7008152300001000000♠152,300,001 | 7008155300000000000♠155,300,000 | gpos | 50 |
| 3 | q | 25.31 | 9162 | 9264 | 7008155300001000000♠155,300,001 | 7008157300000000000♠157,300,000 | gneg | |
| 3 | q | 25.32 | 9264 | 9366 | 7008157300001000000♠157,300,001 | 7008159300000000000♠159,300,000 | gpos | 50 |
| 3 | q | 25.33 | 9366 | 9453 | 7008159300001000000♠159,300,001 | 7008161000000000000♠161,000,000 | gneg | |
| 3 | q | 26.1 | 9453 | 9803 | 7008161000001000000♠161,000,001 | 7008167900000000000♠167,900,000 | gpos | 100 |
| 3 | q | 26.2 | 9803 | 9949 | 7008167900001000000♠167,900,001 | 7008171200000000000♠171,200,000 | gneg | |
| 3 | q | 26.31 | 9949 | 10183 | 7008171200001000000♠171,200,001 | 7008176000000000000♠176,000,000 | gpos | 75 |
| 3 | q | 26.32 | 10183 | 10329 | 7008176000001000000♠176,000,001 | 7008179300000000000♠179,300,000 | gneg | |
| 3 | q | 26.33 | 10329 | 10489 | 7008179300001000000♠179,300,001 | 7008183000000000000♠183,000,000 | gpos | 75 |
| 3 | q | 27.1 | 10489 | 10620 | 7008183000001000000♠183,000,001 | 7008184800000000000♠184,800,000 | gneg | |
| 3 | q | 27.2 | 10620 | 10737 | 7008184800001000000♠184,800,001 | 7008186300000000000♠186,300,000 | gpos | 25 |
| 3 | q | 27.3 | 10737 | 10883 | 7008186300001000000♠186,300,001 | 7008188200000000000♠188,200,000 | gneg | |
| 3 | q | 28 | 10883 | 11175 | 7008188200001000000♠188,200,001 | 7008192600000000000♠192,600,000 | gpos | 75 |
| 3 | q | 29 | 11175 | 11700 | 7008192600001000000♠192,600,001 | 7008198295559000000♠198,295,559 | gneg |
References
^ "Human Genome Assembly GRCh38 – Genome Reference Consortium". National Center for Biotechnology Information. 2013-12-24. Retrieved 2017-03-04..mw-parser-output cite.citation{font-style:inherit}.mw-parser-output .citation q{quotes:"""""""'""'"}.mw-parser-output .citation .cs1-lock-free a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/6/65/Lock-green.svg/9px-Lock-green.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-limited a,.mw-parser-output .citation .cs1-lock-registration a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/d/d6/Lock-gray-alt-2.svg/9px-Lock-gray-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .citation .cs1-lock-subscription a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/a/aa/Lock-red-alt-2.svg/9px-Lock-red-alt-2.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration{color:#555}.mw-parser-output .cs1-subscription span,.mw-parser-output .cs1-registration span{border-bottom:1px dotted;cursor:help}.mw-parser-output .cs1-ws-icon a{background:url("//upload.wikimedia.org/wikipedia/commons/thumb/4/4c/Wikisource-logo.svg/12px-Wikisource-logo.svg.png")no-repeat;background-position:right .1em center}.mw-parser-output code.cs1-code{color:inherit;background:inherit;border:inherit;padding:inherit}.mw-parser-output .cs1-hidden-error{display:none;font-size:100%}.mw-parser-output .cs1-visible-error{font-size:100%}.mw-parser-output .cs1-maint{display:none;color:#33aa33;margin-left:0.3em}.mw-parser-output .cs1-subscription,.mw-parser-output .cs1-registration,.mw-parser-output .cs1-format{font-size:95%}.mw-parser-output .cs1-kern-left,.mw-parser-output .cs1-kern-wl-left{padding-left:0.2em}.mw-parser-output .cs1-kern-right,.mw-parser-output .cs1-kern-wl-right{padding-right:0.2em}
^ ab "Search results – 3[CHR] AND "Homo sapiens"[Organism] AND ("has ccds"[Properties] AND alive[prop]) – Gene". NCBI. CCDS Release 20 for Homo sapiens. 2016-09-08. Retrieved 2017-05-28.
^ Tom Strachan; Andrew Read (2 April 2010). Human Molecular Genetics. Garland Science. p. 45. ISBN 978-1-136-84407-2.
^ abc Genome Decoration Page, NCBI. Ideogram data for Homo sapience (850 bphs, Assembly GRCh38.p3). Last update 2014-06-03. Retrieved 2017-04-26.
^ Pertea M, Salzberg SL (2010). "Between a chicken and a grape: estimating the number of human genes". Genome Biol. 11 (5): 206. doi:10.1186/gb-2010-11-5-206. PMC 2898077. PMID 20441615.
^ "Statistics & Downloads for chromosome 3". HUGO Gene Nomenclature Committee. 2017-05-12. Retrieved 2017-05-19.
^ "Chromosome 3: Chromosome summary – Homo sapiens". Ensembl Release 88. 2017-03-29. Retrieved 2017-05-19.
^ "Human chromosome 3: entries, gene names and cross-references to MIM". UniProt. 2018-02-28. Retrieved 2018-03-16.
^ "Search results – 3[CHR] AND "Homo sapiens"[Organism] AND ("genetype protein coding"[Properties] AND alive[prop]) – Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
^ "Search results – 3[CHR] AND "Homo sapiens"[Organism] AND ( ("genetype miscrna"[Properties] OR "genetype ncrna"[Properties] OR "genetype rrna"[Properties] OR "genetype trna"[Properties] OR "genetype scrna"[Properties] OR "genetype snrna"[Properties] OR "genetype snorna"[Properties]) NOT "genetype protein coding"[Properties] AND alive[prop]) – Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
^ "Search results – 3[CHR] AND "Homo sapiens"[Organism] AND ("genetype pseudo"[Properties] AND alive[prop]) – Gene". NCBI. 2017-05-19. Retrieved 2017-05-20.
^ CRBN cereblon [Homo sapiens (human)] - Gene - NCBI
^ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (400 bphs, Assembly GRCh38.p3). Last update 2014-03-04. Retrieved 2017-04-26.
^ Genome Decoration Page, NCBI. Ideogram data for Homo sapience (550 bphs, Assembly GRCh38.p3). Last update 2015-08-11. Retrieved 2017-04-26.
^ International Standing Committee on Human Cytogenetic Nomenclature (2013). ISCN 2013: An International System for Human Cytogenetic Nomenclature (2013). Karger Medical and Scientific Publishers. ISBN 978-3-318-02253-7.
^ Sethakulvichai, W.; Manitpornsut, S.; Wiboonrat, M.; Lilakiatsakun, W.; Assawamakin, A.; Tongsima, S. (2012). "Estimation of band level resolutions of human chromosome images" (PDF). In Computer Science and Software Engineering (JCSSE), 2012 International Joint Conference on: 276–282. doi:10.1109/JCSSE.2012.6261965.
^ "p": Short arm; "q": Long arm.
^ For cytogenetic banding nomenclature, see article locus.
^ ab These values (ISCN start/stop) are based on the length of bands/ideograms from the ISCN book, An International System for Human Cytogenetic Nomenclature (2013). Arbitrary unit.
^ gpos: Region which is positively stained by G banding, generally AT-rich and gene poor; gneg: Region which is negatively stained by G banding, generally CG-rich and gene rich; acen Centromere. var: Variable region; stalk: Stalk.
External links
| Wikimedia Commons has media related to Human chromosome 3. |
National Institutes of Health. "Chromosome 3". Genetics Home Reference. Retrieved 2017-05-06.
"Chromosome 3". Human Genome Project Information Archive 1990–2003. Retrieved 2017-05-06.



Comments
Post a Comment